RESEARCH FOCUS
Plants cannot move, so they need to be able to adapt quickly to a changing environment. The Cell Cycle Lab aims to understand how plants adjust their growth at the cellular level, more precisely focusing on the process of cell division. Identifying the mechanisms that control cell cycle progression in response to environmental cues may help in the development of climate-resilient crops and innovative crop management practices.
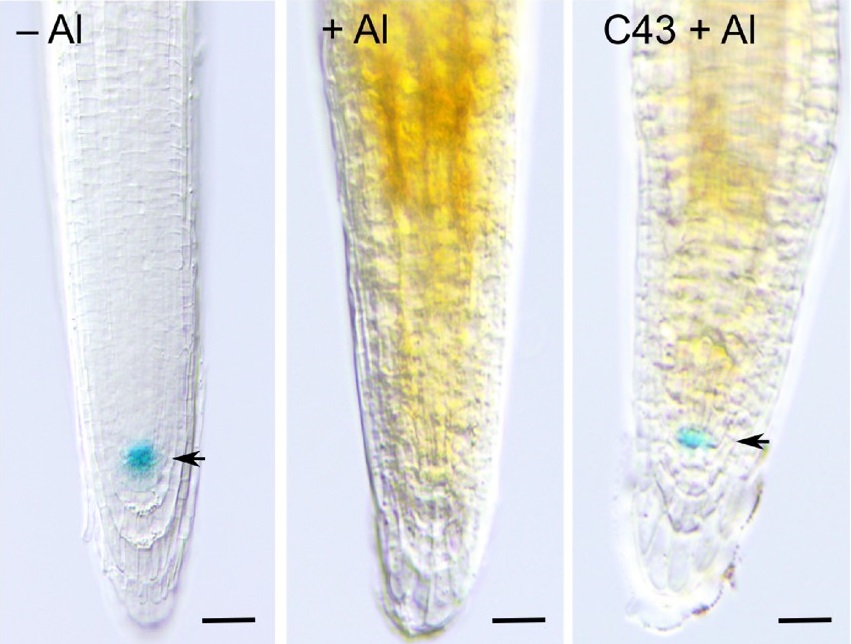
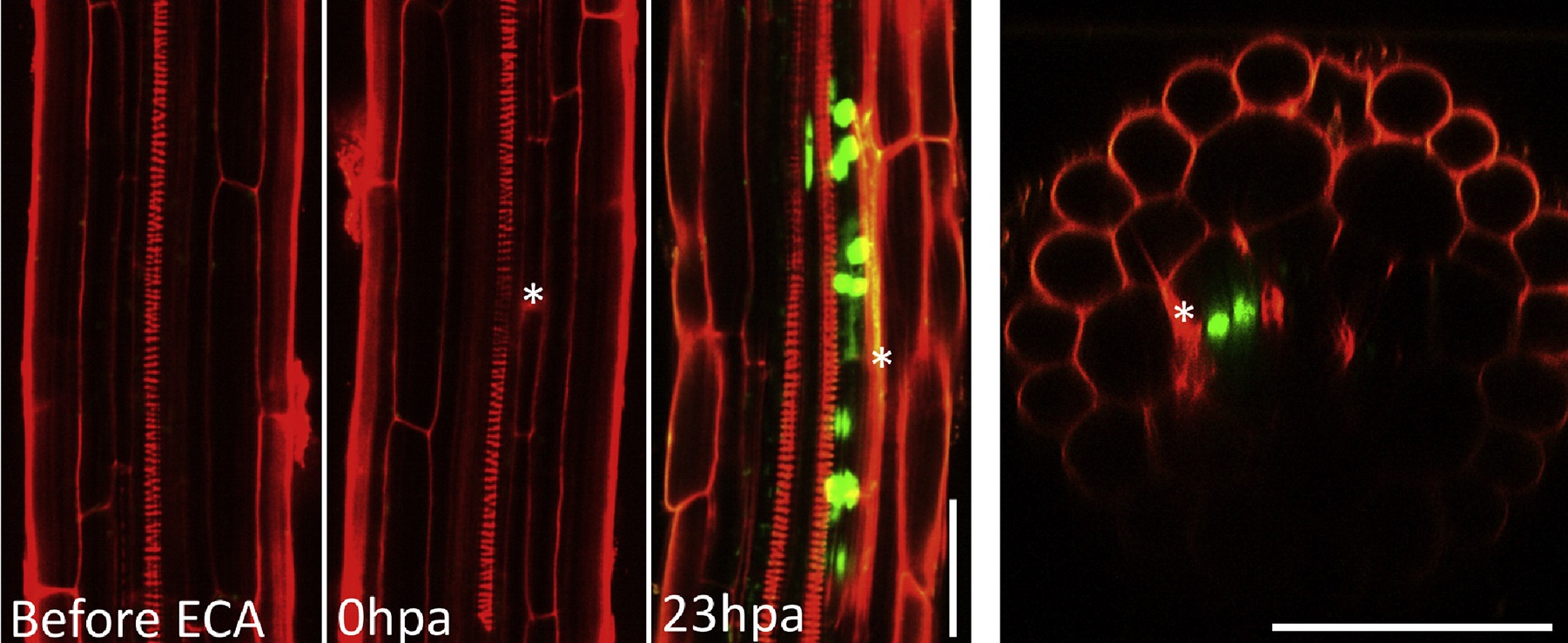
One line of research focusses on wound-induced regeneration. Plants have an unparalleled ability to regenerate, allowing them to survive severe injury, inflicted by herbivory attack or harsh weather conditions. Previously, using the model plant Arabidopsis, we have identified a key molecular component that controls stem cell replenishment after apoptosis, namely a transcription factor nominated ERF115. Even as little as a single dying cell rapidly activates ERF115 in its surrounding cells. Subsequently, it stimulates cells to divide by increasing auxin sensitivity, thereby replacing the damaged cells. Based on these observations, we aim to understand the pathways that activate plant regeneration after wounding and to map the signaling cascades that operate both upstream and downstream of ERF115. Eventually, we aim to generate knowledge that could help the regeneration process of recalcitrant plants.
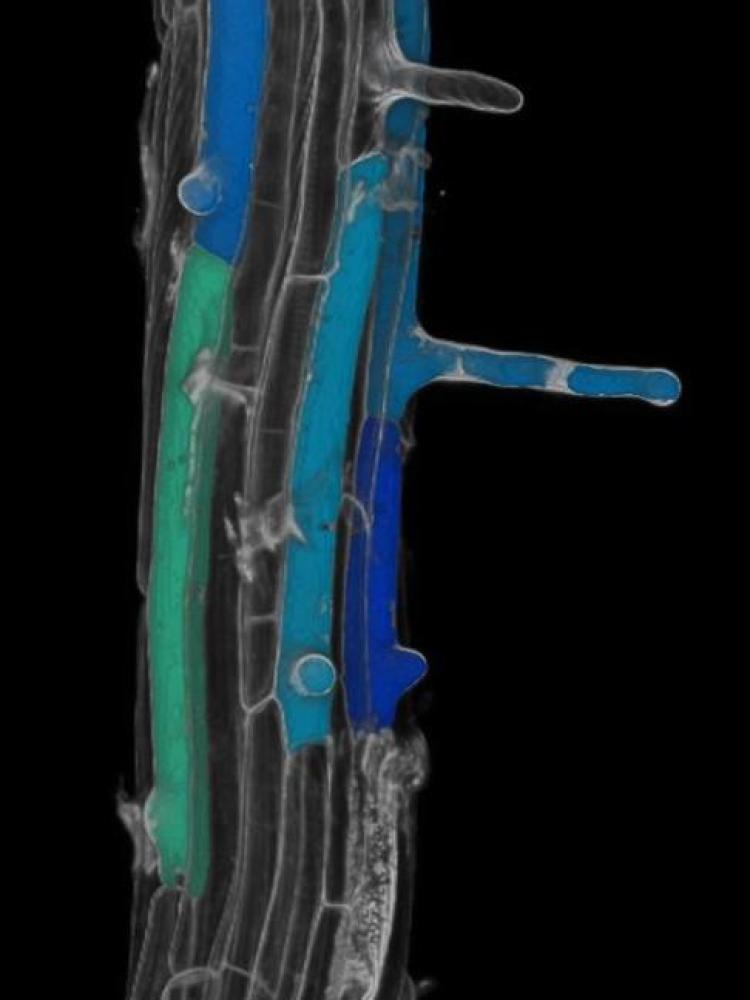
A second line of research involves the DNA damage response (DDR) pathway. The DDR pathway is a highly controlled gene regulatory network, critical for maintaining genome integrity, especially under extreme environmental conditions. To date, plant DDR research has largely focused on the model plant Arabidopsis thaliana. As a result, our knowledge of DNA repair mechanisms in other plant species is limited. Using the revolutionary CRISPR/Cas9 genome editing technology, we have generated DDR signaling mutants in maize and Marchantia. These model systems will be used to study the evolution of the DDR pathway in safe-guarding genome stability. Ultimately, the gained knowledge will be used to create DNA damage-tolerant varieties to increase plant resilience to climate change.
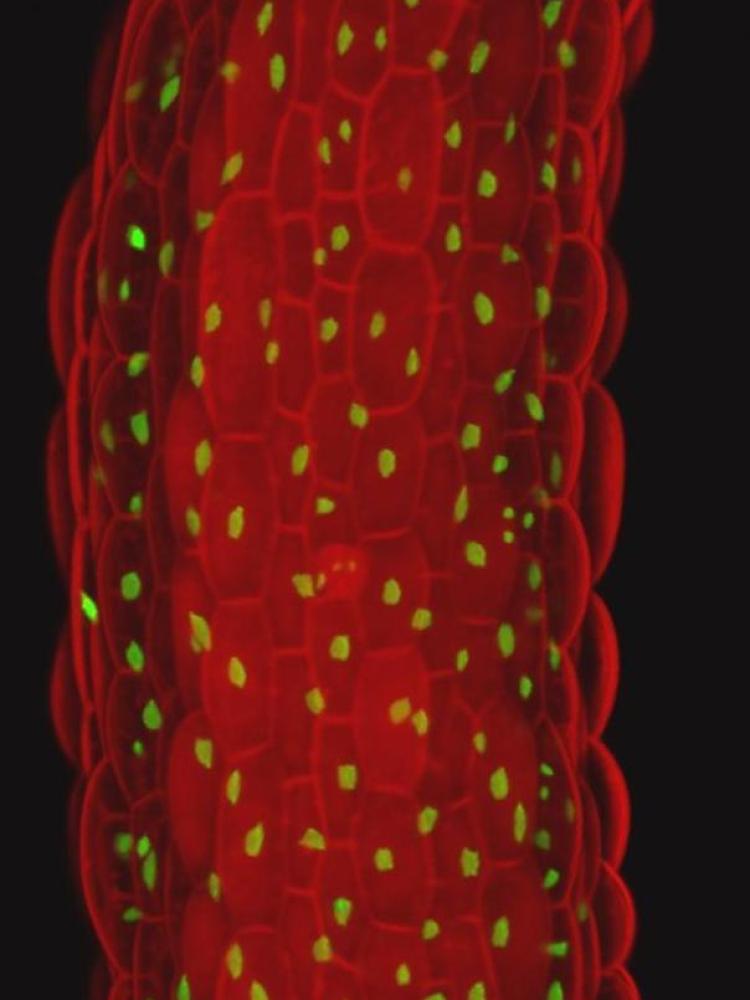
In a third line of research, we are studying a specific class of cell cycle inhibitors called SIAMESE/SIAMESE-RELATED proteins (SIM/SMRs). These proliferation arresting proteins respond specifically to environmental conditions and control developmental transitions, such as the differentiation of stomatal lineage cells into pavement cells. Using the SIM/SMRs genes as a handle, we aim to understand how cell cycle exit is linked to cell differentiation in both Arabidopsis and maize.
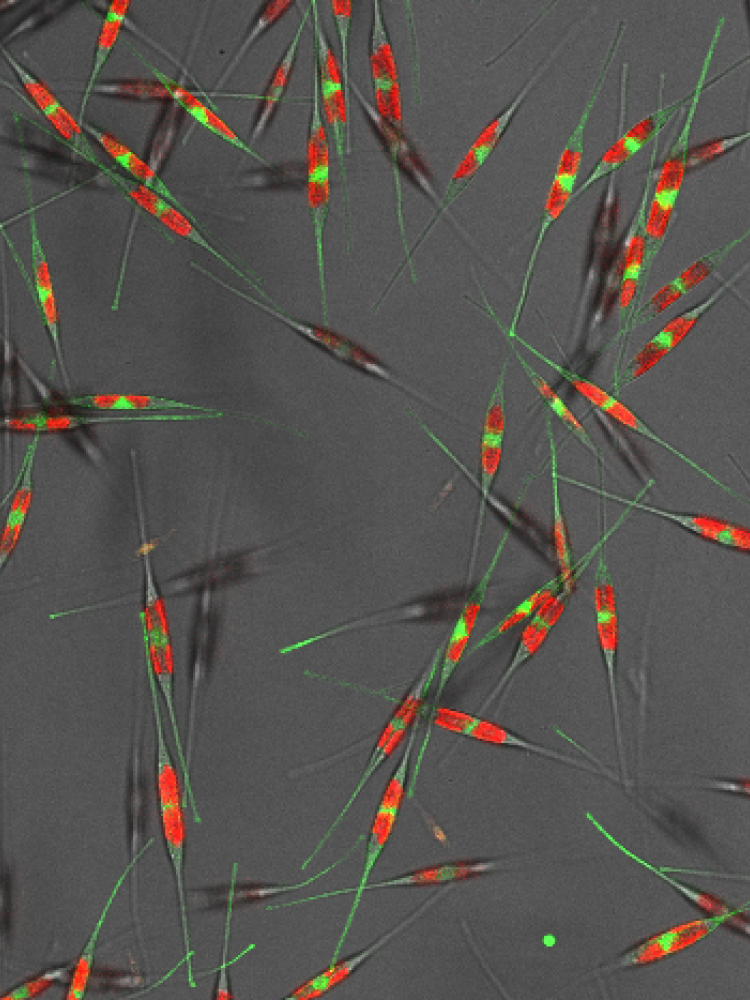
Diatoms are ideal model organisms to study the effects of environmental factors on cell division because they live under variable conditions that require them to adapt to rapid and intense changes in temperature, nutrient availability and light intensity. Previously, the team discovered the mechanism by which diatoms coordinate cell cycle onset with light availability and identified mitotic recombination as a putative stress adaptation mechanism. In follow-up work, the observed mitotic recombination rates will be used as a tool to screen and identify novel genes that allow diatoms to adapt to environmental conditions. In addition, we aim to map the role of mitotic recombination in the diatom's potential to survive harsh conditions.