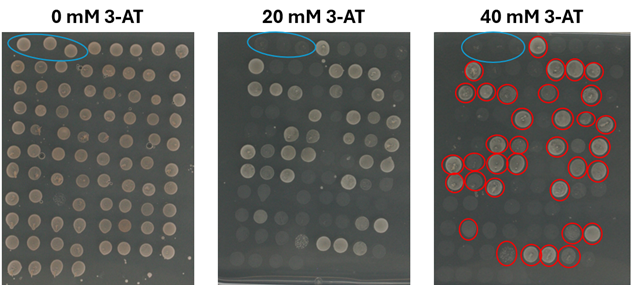
Because plants are often exposed to adverse conditions, they have evolved complex mechanisms to modulate their cell cycle and force them to stop growing temporarily when conditions become too harsh. The main drivers of the cell cycle are cyclin-dependent kinases (CDKs). These CDKs are negatively regulated by the binding of small inhibitory proteins known as CDK inhibitors (CKIs). Among these, the INHIBITORS SIAMESE/SIAMESE-RELATED proteins (SIM/SMRs) are of particular interest, as they have been shown to respond to environmental conditions and control developmental processes. Recently, we have shown that Arabidopsis SMR1 (also called LGO for LOSS OF GIANT CELLS FROM ORGANS) terminates the self-renewal potential of stomatal lineage ground cells, preventing them from re-entering the cell cycle. In this way, SMR1 triggers the differentiation of stomatal lineage cells into pavement cells. Similarly, we have previously shown that SMR4, SMR5 and SMR7 respond to DNA damage and oxidative stress, probably by arresting the cell cycle under conditions that can cause loss of genome integrity. Consistently, smr5 knockout plants show less reduction in leaf epidermal cell number compared to control plants when grown under oxidative stress conditions. Based on these observations and using a variety of genetic and computational tools, we aim to identify upstream regulators of various SIM/SMR genes. In parallel, we aim to investigate their contribution to growth and stress response in crop species.